9.4 Graph Theory - Consensus
convergence of agents' behavior
1Reading¶
Material related to this page can be found in
2Learning Objectives¶
By the end of this page, you should know:
- what is a consensus protocol
- the consensus theorem
- how the eigenvalues/eigenvectors of the Laplacian matrix relate to consensus in a graph
3Consensus Protocols¶
Consider a collection of agents that communicate along a set of undirected links described by a graph . Each agent has state , with initial value , and together, they wish to determine the average of the initial states .
The agents implement the following consensus protocol:
where is the average of the states of the neighbors of agent . This is equivalent to the first-order homogeneous linear ordinary differential equation:
Based on our previous analysis of such systems, we know that the solution to (AVG) is given by
where , , are the eigenvalue/eigenvector pairs of the negative graph Laplacian . Thus, the behavior of the consensus system (AVG) is determined by the spectrum of . We will spend the rest of this lecture on understanding the following theorem:
This result is extremely intuitive! It says that so long as the information at one node can eventually reach every other node in the graph, then we can achieve consensus via the protocol (AVG). Let’s try to understand why. As in the previous lecture, we order the eigenvalues of in decreasing order: .
Our first observation is that , is an eigenvalue/eigenvector pair for . This follows from the fact that each row of sums to 0, and so:
A fact that we’ll show is true later is that the eigenvalues of are all nonnegative, and thus we know that for the eigenvalues of . As such, we know that is the largest eigenvalue of : hence we label them , .
Next, we recall that for an undirected graph, the Laplacian is symmetric, and hence is diagonalized by an orthonormal eigenbasis , where is an orthogonal matrix composed of orthonormal eigenvectors of , and . Although we do not know , we know that .
We can therefore rewrite (SOL) as:
where now we can compute by solving , as is an orthogonal matrix.
Let’s focus on computing :
Plugging this back into (5), we get:
This is very exciting! We have shown that the solution to (AVG) is composed of a sum of the final consensus state and exponential functions , , evolving in the subspace orthogonal to the consensus direction . Thus, if we can show that , we will have established our result.
To establish this result, we start by stating a widely used theorem for bounding localizing eigenvalues.
Let’s apply this theorem to a graph Laplacian . The diagonal elements of are given by , the out-degree of node . Further, the radii as well, as if node is connected to node , and 0 otherwise. Therefore, for row , we have the following Gershgorin intervals:
These are intervals of the form , and therefore the union , where is the maximal out degree of a node in the graph. Taking the negative of everything, we conclude that .
This tells us that for for the eigenvalues of . This is almost what we wanted. We still need to show that only and that . To answer this question, we rely on the following proposition:
Unfortunately proving this result would take us too far astray. Instead, we highlight the intuitive nature of the result in terms of the consensus system (AVG). This proposition tells us that if the communication graph is strongly connected, i.e., if everyone’s information eventually reaches everyone, then at a rate governed by the slowest decaying node .
In contrast, suppose the graph is disconnected, and consists of the disjoint union of two connected graphs and , i.e., and and . Then if we run the consensus protocol (AVG) on , the system effectively decouples into two parallel systems, each evolving on their own graph and blissfully unaware of the other:
Here we use to denote the state of agents in , with Laplacian , and similarly for . By the above discussion, if and are both strongly connected, then , and is an eigenvalue/vector pair for each graph.
If we now consider the joint graph composed of the two disjoint graphs and , we can immediately see how to change our consensus protocol: will evolve as it did before.
To see how this manifests in the algebraic multiplicity of the 0 eigenvalue of , note that for the composite system with state , we have the consensus dynamics:
which has with and with so that:
This is of course expected, as all we have done is rewrite (13) using block vectors and matrices --- we have not changed anything about the consensus protocol.
4Python Break: Determining if a Graph is Connected!¶
In this section, let’s look at an efficient algorithm to check if an (undirected) graph is connected, i.e., has exactly one connected component. Of course, we showed above that the zero eigenvalue has algebraic multiplicity one in the Laplacian of if and only if is connected, which gives us one avenue of attack: to find the eigenvalues of .
In this section, we will explore how a much simpler class of algorithms, known as graph traversal algorithms, can be used to find the connected components of a graph (and count how many there are). The particular class of algorithm we will look at are called tree-based graph traversal algorithms. At a high level, the algorithm starts maintains a set of visited vertices, and at each step, chooses an unvisited vertex that is adjacent to a visited vertex and visits it; the algorithm starts at an arbitrary vertex. A visual can be found below:

From left to right, top to bottom, we see that at each iteration, we choose a vertex (marked in blue) that is adjacent to a green vertex and make it green (visit it). If we start this procedure from a vector , then this procedure will in fact visit all vertices in the connected component of . Try to convince yourself of this fact! To start, recall that is in the connected component of if and only if there is a path from to in the graph.
Below, we have an implementation of this procedure in Python:
import random
from random import randrange
random.seed(6928)
class Graph:
def __init__(self, n):
self.n_vertices = n
self.neighbors = [set() for _ in range(n)]
def add_edge(self, i, j):
self.neighbors[i].add(j)
self.neighbors[j].add(i)
def n_edges(self):
return int(sum([len(s) for s in self.neighbors]) / 2)
def print_edges(self):
for v in range(self.n_vertices):
for neighbor in self.neighbors[v]:
if neighbor > v:
print('(' + str(v) + ', ' + str(neighbor) + ')')
def connected_component(self, v):
cc = set()
visited = [False for _ in range(self.n_vertices)]
frontier = [v]
while len(frontier) > 0:
index = randrange(len(frontier))
v = frontier.pop(index)
if visited[v]:
continue
visited[v] = True
cc.add(v)
for neighbor in self.neighbors[v]:
frontier.append(neighbor)
return cc
g = Graph(5)
g.add_edge(0, 1)
g.add_edge(1, 2)
g.add_edge(2, 0)
g.add_edge(3, 4)
print('Edges of g:')
g.print_edges()
print('Connected component of 0:', g.connected_component(0))
print('Connected component of 1:', g.connected_component(1))
print('Connected component of 2:', g.connected_component(2))
print('Connected component of 3:', g.connected_component(3))
print('Connected component of 4:', g.connected_component(4))
print()
h = Graph(5)
h.add_edge(0, 1)
h.add_edge(1, 2)
h.add_edge(2, 0)
h.add_edge(3, 4)
h.add_edge(0, 4)
print('Edges of h:')
h.print_edges()
print('Connected component of 0:', h.connected_component(0))
print('Connected component of 1:', h.connected_component(1))
print('Connected component of 2:', h.connected_component(2))
print('Connected component of 3:', h.connected_component(3))
print('Connected component of 4:', h.connected_component(4))Edges of g:
(0, 1)
(0, 2)
(1, 2)
(3, 4)
Connected component of 0: {0, 1, 2}
Connected component of 1: {0, 1, 2}
Connected component of 2: {0, 1, 2}
Connected component of 3: {3, 4}
Connected component of 4: {3, 4}
Edges of h:
(0, 1)
(0, 2)
(0, 4)
(1, 2)
(3, 4)
Connected component of 0: {0, 1, 2, 3, 4}
Connected component of 1: {0, 1, 2, 3, 4}
Connected component of 2: {0, 1, 2, 3, 4}
Connected component of 3: {0, 1, 2, 3, 4}
Connected component of 4: {0, 1, 2, 3, 4}
Above, we define two graphs g and h and find the connected component corresponding to each vertex in the two graphs using the connected_components function. Note that g has two connected components, {0, 1, 2} and {3, 4} whereas h has only one connected component {0, 1, 2, 3, 4}, meaning that h is connected and g is not.
Let’s understand how this algorithm implements the procedure we described above. At a high level, at each iteration of the while loop, the cc set will contain all the vertices we have already visited (the visited array will also indicate which vertices we have already visited), and the frontier list will contain all the unvisited vertices adjacent to any vertex we have already visited. At each iteration of the while loop, we pick a random vertex in the frontier list and visit it, and then repeat until there are no more vertices to explore (i.e., we have explored the entire connected component). At the end of this procedure, the cc set will contain all the vertices in our connected component!
To conclude, we have a few remarks about the way we choose vertices from the frontier list. Currently, we randomly select vertices from this list, but we can instead use an arbitrary rule to choose which vertex instead. For example:
If we choose the vertex in
frontierwhich was added earliest (out of all vertices which are infrontier), then this algorithm is called breadth-first search, or BFS.If we choose the vertex in
frontierwhich was most recently added, then this algorithm is called depth-first search, or DFS.
A visual of BFS versus DFS is below:
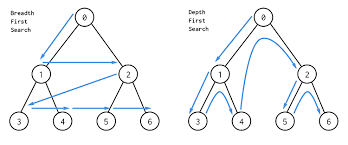
If you take a computer science class, you’ll definitely see these very commonly used algorithms in a lot more detail!